Interactive browser, part 2: scalable clustering
Building clusterings into a front-end loop
- Intro: interactive clustering of data
- Loading the previous browser
- A heatmap to display the features
- Co-clustering to group the selected data
- Displaying and viewing the resulting heatmap
- Summary: interactive clustering
- References
Intro: interactive clustering of data
Previous posts in the series: [Part 1]
Interactive browsers have started to enter wider usage in various biochemical fields which observe extremely high-dimensional and structured data on a regular basis. I recently wrote about building the foundations of such a browser for visualizing chemical space, in which the user can define subsets of chemicals interactively.
Browsers like this need to perform general learning tasks without making unnecessary assumptions. The potential of this workflow can be realized through interactive algorithms driven by user selections. The theme of interactive computation is the focus of this post series. I'm going to explore it in this post, by demonstrating how to add a heatmap, and augment it to group subsets of chemicals based on their fingerprints.
Loading the previous browser
I'm first loading the previously created interactive browser, as described in the previous post in the series. That post covers the basics of how to build an interactive interface with subselection capabilities. The code to run the browser is imported from the relevant file produced in that previous post.
import requests
url = 'https://raw.githubusercontent.com/b-akshay/blog-tools/main/interactive-browser/part-1/scatter_chemviz.py'
r = requests.get(url)
# make sure your filename is the same as how you want to import
with open('VE_browser.py', 'w') as f:
f.write(r.text)
# now we can import
from VE_browser import *
A heatmap to display the features
I'm going to add another panel to the interface, with profound implications. This is a heatmap that displays feature-level values for every observation, i.e. it displays the "raw data" -- in this case, the values of individual fingerprints for a single molecule. It allows the user to define subgroups of the data based on their representations directly, rather than based on some derived 2D scatterplots.
Selecting features to view
In this case, we could use my original featurization of 2048 Morgan (extended connectivity) fingerprint bits for this chemical data.
The heatmap only takes up less than half the screen horizontally - a few hundred pixels. So there are clearly too many features to view individually - more than the number of pixels displaying the heatmap. This is quite common in data visualization, and it is crucially important which features are selected to view.
As a first pass, I've implemented a simple function that selects the maximum-variance features in the data.
def interesting_feat_ndces(fit_data, num_feats_todisplay=500):
num_feats_todisplay = min(fit_data.shape[1], num_feats_todisplay)
if ((fit_data is None) or
(np.prod(fit_data.shape) == 0)
):
return np.arange(num_feats_todisplay)
feat_ndces = np.argsort(np.std(fit_data, axis=0))[::-1][:num_feats_todisplay]
return feat_ndces
This runs at interactive speeds, and provides a guardrail against rendering astronomically many features.
Timing
What do I mean by "interactive speeds"? Running it on this data (>2K features) takes about 0.02s on my laptop. This is far below the ~1 second needed to perceptually throw the user out of the feeling of interactivity, a hard limit on our interactive designs that I've written about recently. Timing this feature selection step is easy, and good practice.
%timeit interesting_feat_ndces(anndata_all.X)
Displaying metadata
Though it's possible to interactively narrow down to a manageable number of fingerprint bits, they are not easy to individually interpret. So we opt instead to display some descriptors calculated using various human-interpretable RDKit utilities, as I wrote about earlier.
These are already stored as metadata columns in the .obs dataframe; there are 208 such columns we display as a heatmap.
print("Shape of displayed metadata: {}".format(anndata_all.obs.iloc[:, :-8].values.shape))
Co-clustering to group the selected data
This uses a function compute_coclustering which groups the rows and columns, and returns the cluster indices and IDs.
We focus on one operation in particular: ordering the rows and columns of the heatmap to emphasize the internal structure. This can be done with co-clustering -- assigning joint cluster labels to the rows and columns of the heatmap.
An implementation with linear algebra
There are many methods for co-clustering, including work linking it to information theory [Dhillon et al. 2003]. I'll use the most efficient, the spectral partitioning method of [Dhillon 2001]. This is implemented in Scikit-learn. Here's a wrapper around it setting it up for use by the browser.
from sklearn.cluster import SpectralCoclustering
def compute_coclustering(
fit_data,
num_clusters=1,
tol_bicluster=0.005, # sparsity otherwise annoyingly causes underflows w/ sklearn
):
if num_clusters == 1:
num_clusters = min(fit_data.shape[0], 5) # = (working_object.shape[1]//5)
if scipy.sparse.issparse(fit_data):
fit_data = fit_data.toarray()
row_labels, col_labels = cocluster_core_sklearn(fit_data, num_clusters)
return (np.argsort(row_labels), np.argsort(col_labels), row_labels, col_labels)
def cocluster_core_sklearn(
fit_data,
num_clusters,
random_state=0
):
model = SpectralCoclustering(n_clusters=num_clusters, random_state=random_state)
model.fit(fit_data)
return model.row_labels_, model.column_labels_
Timing
This is fast enough to be within the ~0.5-to-1-second perceptual barrier on my laptop, when considering the entire dataset of nearly 6,000 chemicals.
%timeit compute_coclustering(anndata_all.X[:, interesting_feat_ndces(anndata_all.X)])
For more details, read on. Otherwise, to skip to the code that renders the pretty heatmaps, click here.
What it does: reorder the data
Once data are co-clustered, simply arranging them according to their co-cluster labels, so that rows and columns of the same label are in contiguous blocks, reveals latent structure visually in an appealing way.
The scikit-learn documentation has a nice example to convey this idea.
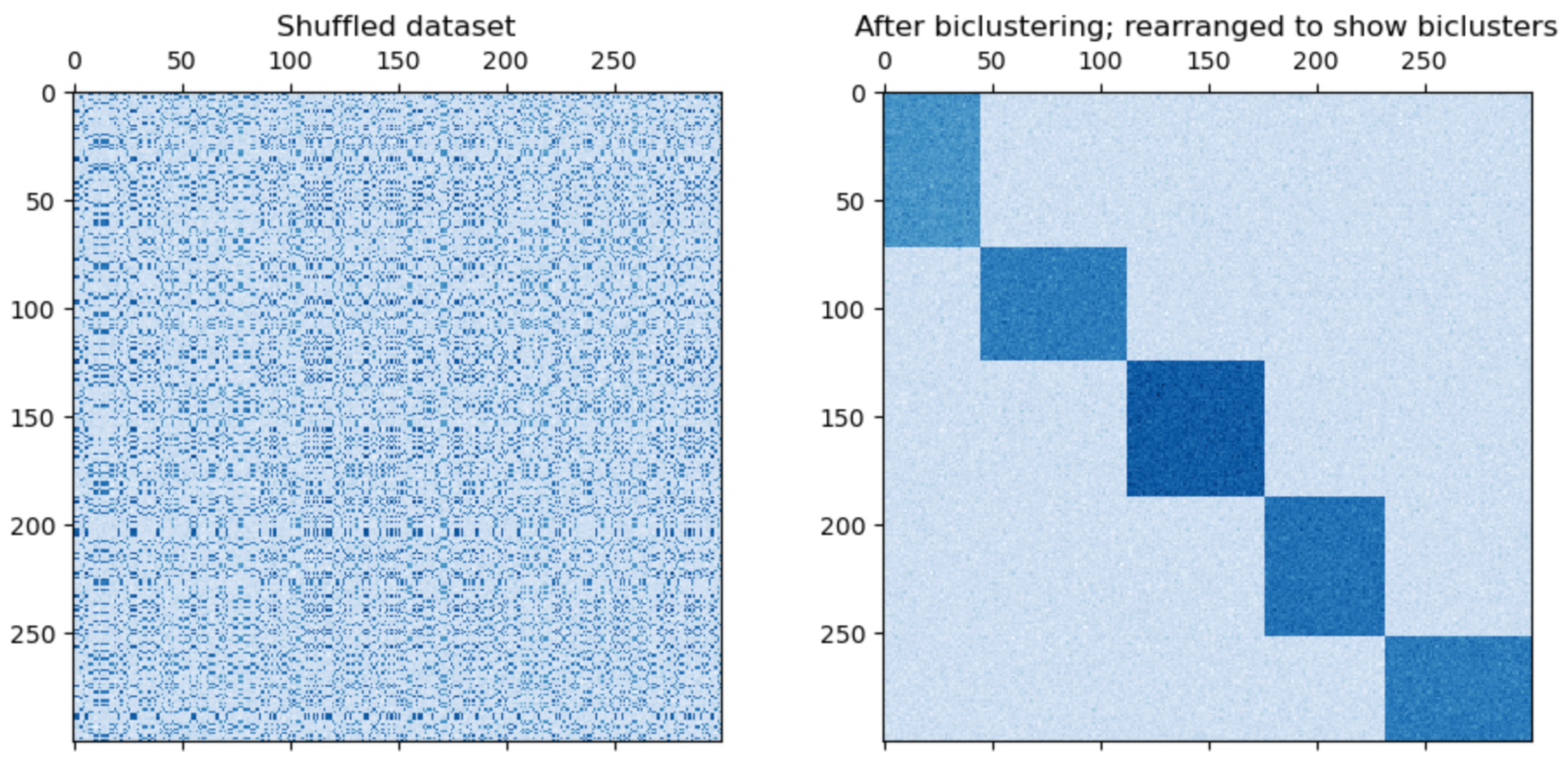
The question is: how does this method find such an ordering efficiently?
Why it's efficient: it relies on linear algebra
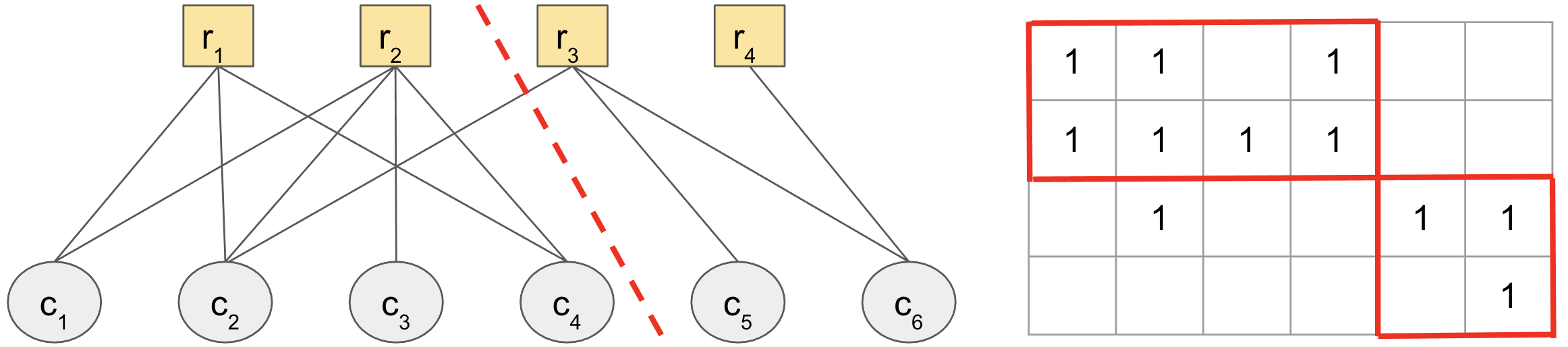
The algorithm is based on the following intuition:
Find a clustering that minimizes entries off the block diagonal.
This is done in a couple of steps:1. Form a bipartite graph from the data matrix, where the rows and columns correspond to vertices, and the entries to edges.2. Find a way of cutting this graph into clusters that minimizes the weight of edges cut.
Step (2) is exactly the same problem faced by spectral clustering, which approximately finds these low-weight balanced graph cuts with linear algebra, by just finding one (or a few) eigenvectors. So it can be efficiently done for the scales of data we're analyzing interactively.
Step (1), making a graph from the data, is where the insight and complexity come in. This makes a bold insight, in considering rows and columns all as first-class entities in the same bipartite graph. Looking at a toy example may help convince us that a minimum cut on this graph (indicated by the dashed red line) is what we want when the data show co-clustered structure.
- Different normalizations of the data will give slightly different results with this method! It pays to try row- and column-wise normalizations.
- More dramatic steps like binarizing the data may also be appropriate for sparse data.
Tip: It does tend to work well when zeroes in the data can be neglected, and higher-magnitude entries are meaningful.Warning: Watch out for all-zero rows/columns! It’s best to remove them.
Displaying and viewing the resulting heatmap
Generating the heatmap in Plotly
Putting it all together, here's some wrapper code for translating all these configuration and style options into Plotly for a heatmap.
${\normalsize \textbf{Details}}$
- The code in
hm_hovertext()creates tooltip text for the heatmap. This is done pretty simply right now, but is where the performance bottleneck is. - The code uses several display configuration options from the
paramsdictionary defined above. The code could be fewer lines, but everyone likes different display configurations! - The main implementation details here arise from the differences in handling discrete and continuous colorscales, which affects subset selection and annotation of points.
- Further details, like the displayed name of the color variable, are set to generic defaults that are exposed in the code and can be changed.
from sklearn.preprocessing import StandardScaler
def hm_hovertext(data, rownames, colnames):
pt_text = []
# First the rows, then the cols
for r in range(data.shape[0]):
pt_text.append(["Observation: {}".format(str(rownames[r])) for k in data[r, :]])
for c in range(data.shape[1]):
pt_text[r][c] += "<br>Feature: {}<br>Value: {}".format(str(colnames[c]), str(round(data[r][c], 3)))
return pt_text
def display_heatmap_cb(
data_df,
color_var,
row_annots=None,
show_legend=False,
col_alphabet=True,
plot_raw=True,
max_cols_heatmap=400,
xaxis_label=True, yaxis_label=True,
scatter_frac_domain=0.10
):
itime = time.time()
if data_df is None or len(data_df.shape) < 2:
return
working_object = data_df
# Identify (interesting) features to plot. Currently: high-variance ones
if working_object.shape[1] > 500:
feat_ndces = interesting_feat_ndces(working_object.values)
working_object = working_object.iloc[:, feat_ndces]
# Here we subsample down to `max_rows_allowed` rows if needed, and make data prettier for printing
max_rows_allowed = 1000
if working_object.shape[0] > max_rows_allowed:
ndxs = np.random.choice(np.arange(working_object.shape[0]), size=max_rows_allowed, replace=False)
working_object = working_object.iloc[ndxs, :]
if row_annots is not None:
row_annots = row_annots[ndxs]
# Spectral coclustering to cluster the heatmap. We always order rows (points) by spectral projection, but cols (features) can have different orderings for different viewing options.
if (working_object.shape[0] > 1):
fit_data = StandardScaler().fit_transform(working_object.values)
ordered_rows, ordered_cols, row_clustIDs, col_clustIDs = compute_coclustering(fit_data)
if row_annots is not None:
ordered_rows = np.lexsort((row_clustIDs, row_annots))
working_object = working_object.iloc[ordered_rows, :]
else:
ordered_cols = np.arange(working_object.shape[1]) # Don't reorder at all
if col_alphabet:
ordered_cols = np.argsort(working_object.columns) # Order columns alphabetically by feature name
working_object = working_object.iloc[:, ordered_cols]
# Finished reordering rows/cols
working_object = working_object.copy()
hm_point_names = np.array(working_object.index)
absc_labels = np.array(working_object.columns)
row_scat_traces = hm_row_scatter(working_object, color_var, hm_point_names)
if not plot_raw:
working_object.values = StandardScaler().fit_transform(working_object.values)
pt_text = hm_hovertext(working_object.values, hm_point_names, absc_labels)
hm_trace = {
'z': working_object.values,
'x': absc_labels,
'customdata': hm_point_names,
'hoverinfo': 'text',
'text': pt_text,
'colorscale': params['colorscale_continuous'],
'colorbar': {
'len': 0.3, 'thickness': 20,
'xanchor': 'left', 'yanchor': 'top',
'title': params['hm_colorvar_name'], 'titleside': 'top', 'ticks': 'outside',
'titlefont': colorbar_font_macro,
'tickfont': colorbar_font_macro
},
'type': 'heatmap'
}
max_magnitude = np.percentile(np.abs(working_object.values), 98) if working_object.shape[0] > 0 else 2
hm_trace['zmin'] = -max_magnitude
hm_trace['zmax'] = max_magnitude
return {
'data': [ hm_trace ] + row_scat_traces,
'layout': {
'xaxis': {
'automargin': True,
'showticklabels': False,
'showgrid': False, 'showline': False, 'zeroline': False, #'visible': False,
#'style': {'display': 'none'},
'domain': [scatter_frac_domain, 1]
},
'yaxis': {
'automargin': True,
'showticklabels': False,
'showgrid': False, 'showline': False, 'zeroline': False, #'visible': False,
#'style': {'display': 'none'}
},
'annotations': [{
'x': 0.5, 'y': 1.10, 'showarrow': False,
'font': { 'family': 'sans-serif', 'size': 15, 'color': params['legend_font_color'] },
'text': 'Features' if xaxis_label else '',
'xref': 'paper', 'yref': 'paper'
},
{
'x': 0.05, 'y': 0.5, 'showarrow': False,
'font': { 'family': 'sans-serif', 'size': 15, 'color': params['legend_font_color'] },
'text': 'Observations' if yaxis_label else '', 'textangle': -90,
'xref': 'paper', 'yref': 'paper'
}
],
'margin': { 'l': 30, 'r': 0, 'b': 0, 't': 70 },
'hovermode': 'closest', 'clickmode': 'event', # https://github.com/plotly/plotly.js/pull/2944/
'uirevision': 'Default dataset',
'legend': style_legend, 'showlegend': show_legend,
'plot_bgcolor': params['bg_color'], 'paper_bgcolor': params['bg_color'],
'xaxis2': {
'showgrid': False, 'showline': False, 'zeroline': False, 'visible': False,
'domain': [0, scatter_frac_domain],
'range': [-1, 0.2]
}
}
}
def hm_row_scatter(data_df, color_var, hm_point_names):
return []
This defines a dummy function hm_row_scatter.
The job of this function is to return a column of scatterplot points alongside the rows to serve as a link between the scatterplot and the heatmap.
Making this link work for significantly better subset selection is a task in itself.
There is also additional code there I've left unexplained for now, involving customizable "row annotations" that can be used to modify the order of what's displayed.
Delving into all this would make our discussion here very long, and is the subject of a subsequent post.
The end product
The code from this notebook is available as a file; setting it up an environment will deploy the app.
There are a couple of interesting stories along the way which merit their own code snippets.
import plotly.graph_objects as go
from IPython.display import HTML
import scanpy as sc, anndata
import time
def custom_colwise_norm_df(data_df):
# Define custom column-wise normalization here.
data_df = data_df.iloc[:, :-8]
for col in data_df.columns:
newvals = np.nan_to_num(data_df[col])
if np.std(newvals) > 0:
newvals = scipy.stats.zscore(newvals)
data_df[col].values[:] = newvals
return data_df
anndata_all = sc.read('approved_drugs.h5')
data_df = anndata_all.obs
data_df = custom_colwise_norm_df(data_df)
# Export scatterplot as HTML.
fig_data = display_heatmap_cb(data_df, 'MolWt')
fig = go.Figure(**fig_data)
# HTML(fig.to_html())
Cumulative timing
Let's time everything so far:
- Feature selection
- Coclustering of the heatmap
- Putting together the Plotly figure
- Rendering the figure (e.g. in HTML)
Recall that for a fully interactive experience, we should keep the overall computation time under ~1s when the user selects a chunk of the scatterplot.
from IPython.display import HTML
%timeit fig_data = display_heatmap_cb(data_df, 'MolWt')
%timeit fig = go.Figure(**fig_data)
%timeit HTML(fig.to_html())
This is not happening with the present design; large heatmaps cannot be rendered without the user feeling like they have to pause their thoughts.
After profiling the code in display_heatmap_cb, it turns out that most of the time is being spent in hm_hovertext.
We leave optimization of this function to a later post.
"""
Update the main heatmap panel.
"""
@app.callback(
Output('main-heatmap', 'figure'),
[Input('landscape-plot', 'selectedData')])
def update_heatmap(
selected_points
):
if (selected_points is not None) and ('points' in selected_points):
selected_IDs = [p['text'].split('<br>')[0] for p in selected_points['points']]
else:
selected_IDs = []
if len(selected_IDs) == 0:
selected_IDs = data_df.index
subsetted_data = data_df.loc[np.array(selected_IDs)]
subsetted_data = custom_colwise_norm_df(subsetted_data)
print(f"Subsetted data: {subsetted_data.shape}")
row_annotations = None
return display_heatmap_cb(
subsetted_data, 'MolWt',
row_annots=row_annotations,
xaxis_label=True, yaxis_label=True
)
Takeaways
Rule of thumb: No more than ~500K entries in the displayed heatmap.
${\normalsize \textbf{Explanation}}$
- Screen capability: A screen has finite resolution, and a row/column of the heatmap takes at least a pixel to intelligibly distinguish. So this will display the maximum possible if e.g. a 1000 x 500 pixel area of the interface being devoted to the heatmap.
- User perception: This also goes back to one of the central principles I talked about in a previous post - all computations should occur in less than a second in order to make a human user feel like the experience is interactive. One of the most intensive types of computations the browser performs is rendering the heatmap and its tens of thousands of individual entries, each with a different tooltip that displays upon hovering. This seems to be a bottleneck at scales around $~10^5$ or more – much more than this cannot comfortably be displayed on most desktop/laptop screens, let alone mobile.
This turns out to be a pretty stringent requirement - even 1000 observations of 500 features each can take a while to render, as we'll see. Such considerations are tied to the framework, and using better heatmap renderers can give significant speedups.
But the datasets we deal with in biochemical sciences are often several orders of magnitude larger. I'll write next about bridging that gap of scale.
Up ahead: scaling up
As the capabilities of this browser grow, we wonder: how much algorithmic functionality can we enable at interactive speeds?
- Next up is a crucial pit stop along this journey, adding several upgrades to the browser's abilities that allow the user to zoom into data-driven subsets.
- After that, we'll open up some corners of the algorithmic toolbox on these subsets, and demonstrate what is possible at these speeds.
One key thing to remember is that the process is already bottlenecked by runtime considerations. Around 10,000 points seem to be all that will visualize at perceptually interactive speeds on a CPU-bound local machine.1 So this sets a practical limit on the size of the datasets passed into algorithms for learning.
In the next posts, we'll look at what algorithmic and visualization functionality we can include to make the user's life easier.
1. However, much more than that is possible using GPUs, with technologies like WebGL.↩
- Dhillon, I.S., Mallela, S., and Modha, D.S. 2003. Information-theoretic co-clustering. Proceedings of the ninth ACM SIGKDD international conference on Knowledge discovery and data mining, 89–98.
- Dhillon, I.S. 2001. Co-clustering documents and words using bipartite spectral graph partitioning. Proceedings of the seventh ACM SIGKDD international conference on Knowledge discovery and data mining, 269–274.