Variable expectations
| Title | Description | Reading Time | |
|---|---|---|---|

|
Navigating published literature with full control | Tracking a field in real time, cheaply | 14 min |
|
Procedurally generating data for tuning technical LLMs | Turning tacit technical knowledge into prediction | 8 min |
|
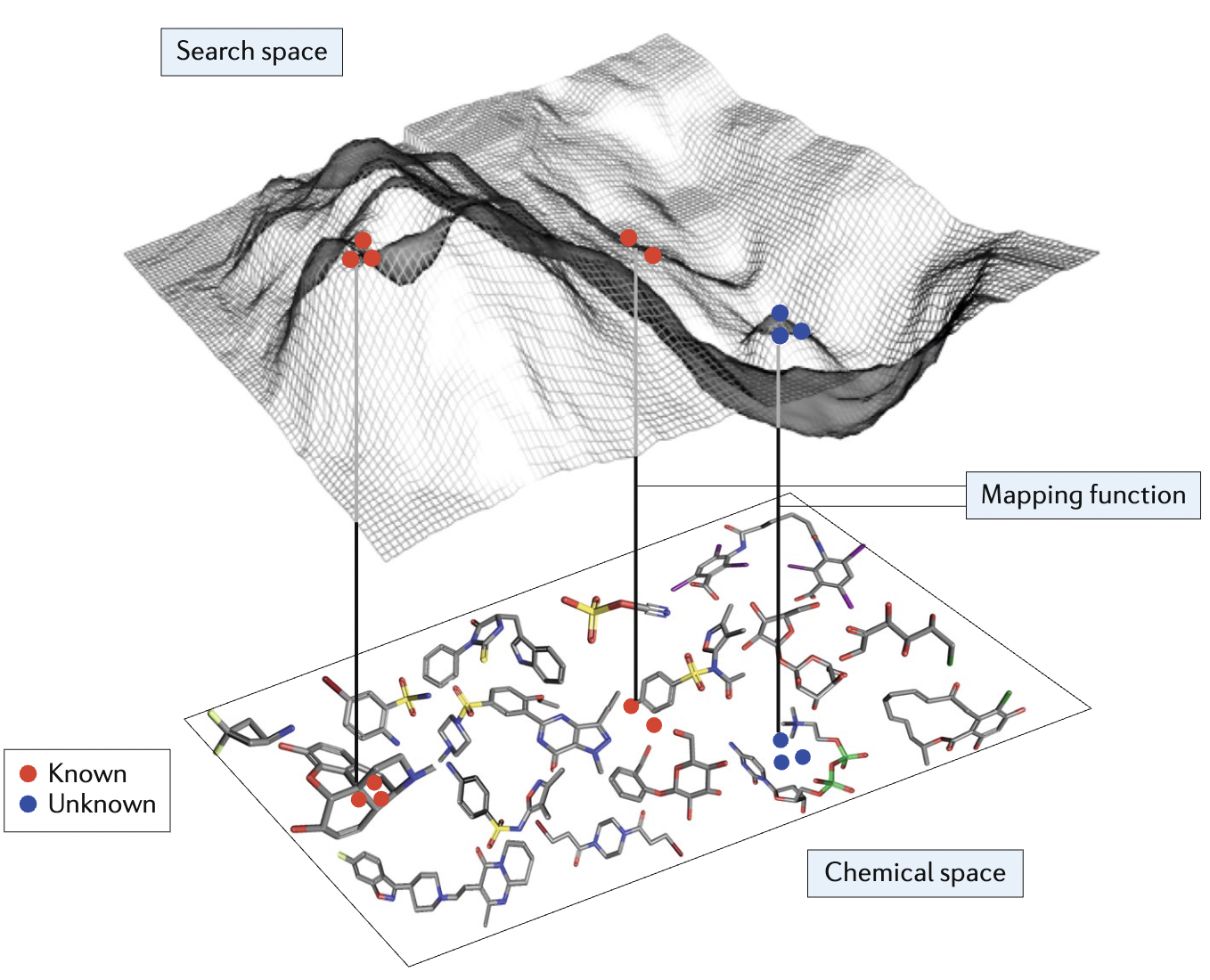
AI in the small-molecule discovery funnel | Virtual spaces and chemical languages | 14 min |
|
Linear algebra in the AI world | Same classics, new applications | 4 min |
|
In silico fragment synthesis with combinatorial chemistry | Revisiting LNP libraries for rational design | 7 min |

|
Combinatorial chemistry and in silico decompositions | Decomposition as inverted synthesis | 10 min |
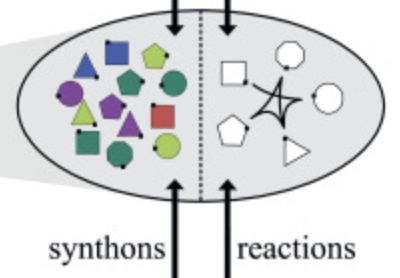
|
Searching too many chemicals to count | Searching trillions of structures on a laptop | 7 min |

|
Embedding ultralarge spaces | Revisiting LNP libraries for rational design | 2 min |

|
Sourcing chemical structures for in silico discovery | Getting ionizable lipid structures from online catalogs | 10 min |

|
Screening too many chemicals to count | Searching trillions of structures on a laptop | 15 min |

|
Decomposition and synthesis with a flexible reaction set | Reaction sets for different in silico ultralarge spaces | 10 min |
|
Making animations and GIFs | Stringing together plots into GIFs | 2 min |
|
Color for an applied data scientist | Embarrassingly useful colormap tidbits | 12 min |

|
Building an interactive data browser | Lightweight, performant exploration of chemical space | 10 min |
|
Graphs as interactive diagrams | Translating between interactive flowcharts and data science | 8 min |

|
Two roads to AI’s future | Superhuman oracles and social intelligences | 15 min |
|
Some useful hashing tricks | Indexing for numbering and obfuscation | 5 min |

|
Visualization tools for science and computation | Increasing capabilities in domain-specific languages | 7 min |
|
Lightweight, interactive QSAR importance plots | A self-contained visualization of atomic contributions to models | 7 min |

|
Interactive browser, part 2 - finding and displaying structure | An interactively clustered heatmap | 8 min |
|
Visualizing in silico chemical decompositions | Ways to view ultralarge chemical spaces | 2 min |
|
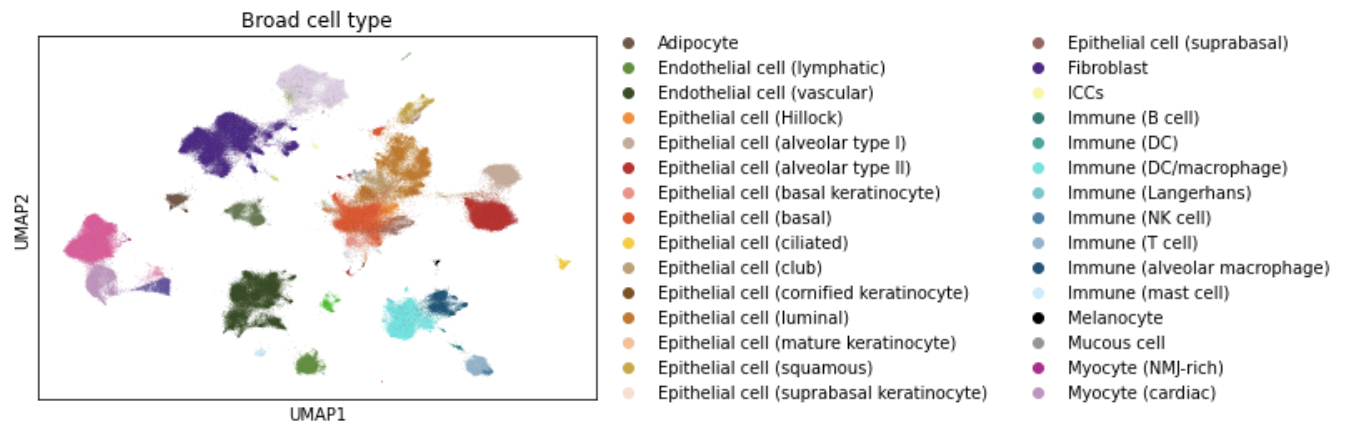
Interactive browser, part 3 - subset selection and scaling | Repurposing for single-cell data | 11 min |

|
Looking up gene function in databases | Cross-referencing many databases at once | 19 min |

|
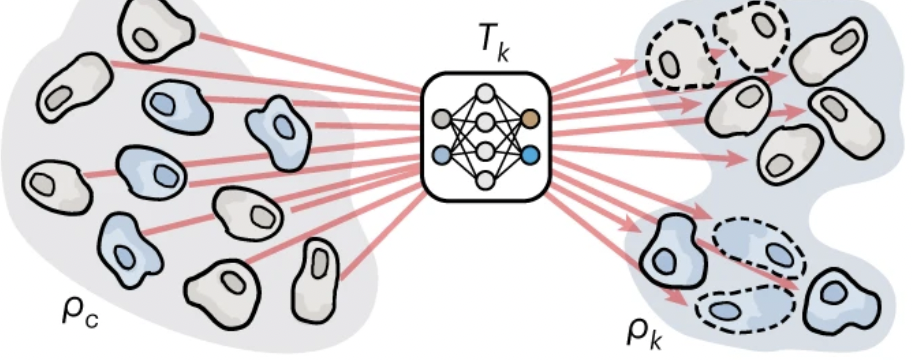
Gene sets from gene function | Deriving gene sets to steer genomic learning | 15 min |
|
Exploring perturbation data in drug discovery | Some trajectories of AI-guided exploration | 5 min |
|
Tree! I am no tree! I am an embedding! | Hierarchies on data | 8 min |
|
Graph sparsification | Removing edges while maintaining connectivity | 13 min |
|
Localized graph descriptions | Learning an interpretable coordinate system for a dataset | 5 min |
|
Laplacian regularization and trend filtering | Using graphs to regularize learning | 9 min |
|
Graph-based semi-supervised prediction | Harmonic analysis for learning on graphs | 8 min |
|
Using graphs to express structure | Efficient, practical tools | 1 min |
|
Calculus on graphs as manifolds | From local change to global order | 15 min |
|
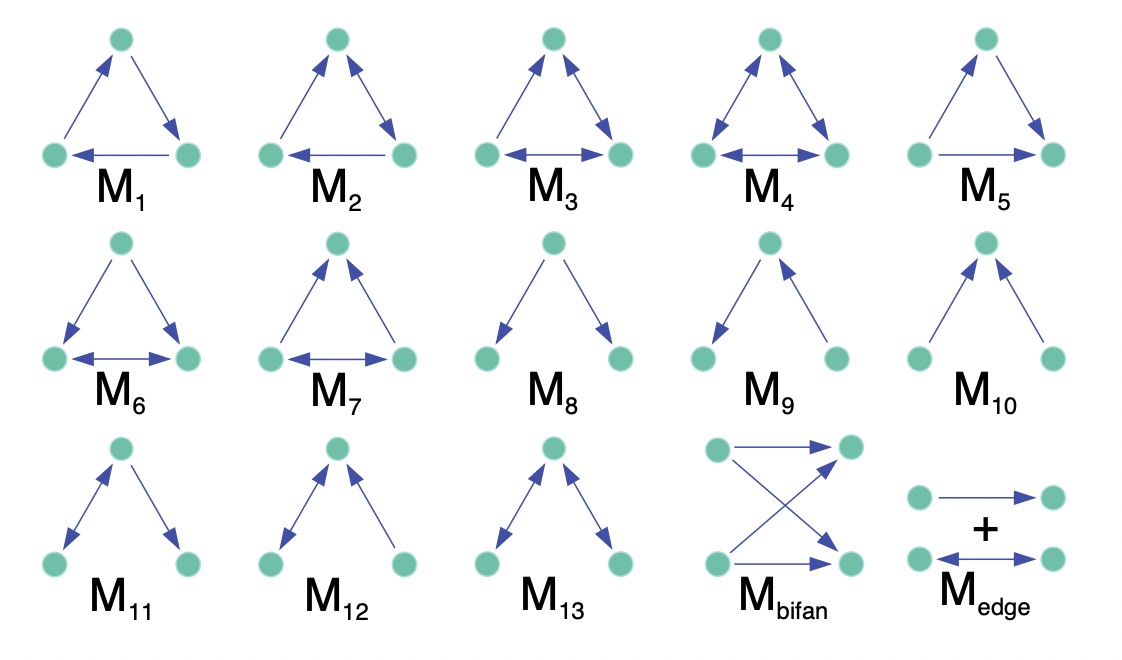
Detecting motifs in networks | Some linear algebra one-liners | 13 min |
|
Global evaluations from two-way comparisons | Quickly learning from pairwise feedback | 10 min |
|
Generating signals from neighborhood graphs | Using graphs as compressed representations | 6 min |
|
How to construct graphs for data analysis | Building neighborhood graphs from datasets | 13 min |
|
Low-rank approximation of matrices and graphs | Efficient, robust, simple, and interpretable | 4 min |

|
The SVD and friends | Breaking down a (sparse) matrix quickly | 12 min |
|
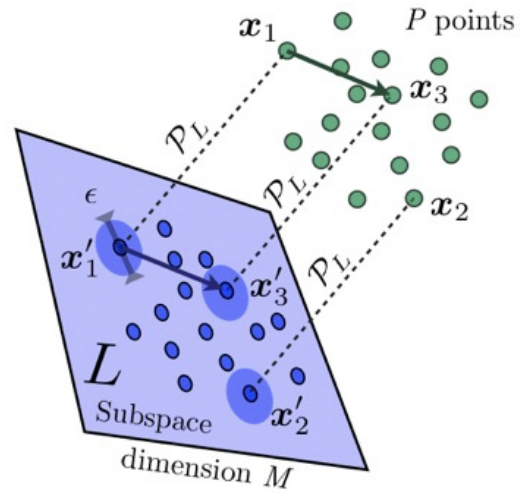
Random projections and dimensionality reduction | Efficient, general applied tools | 10 min |
|
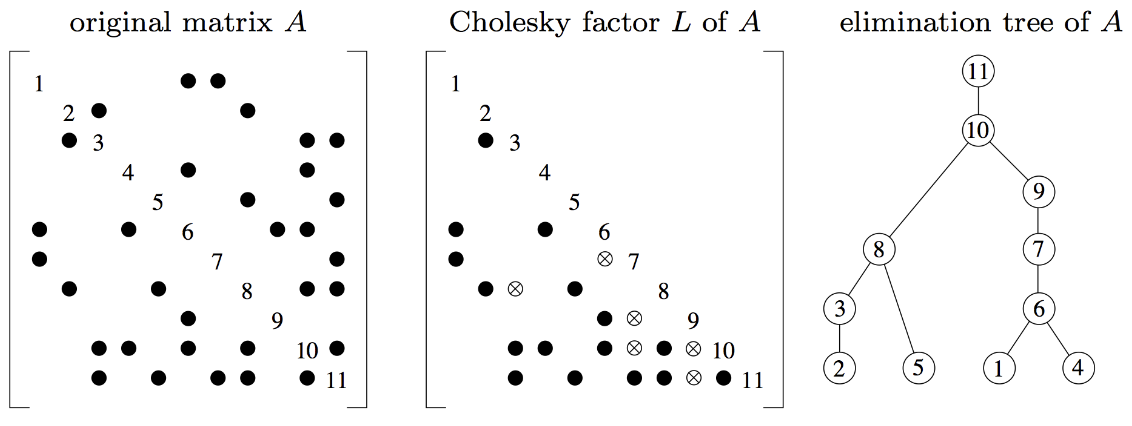
Solving sparse linear equations | Logging the results of Gaussian elimination | 16 min |
|
SVD-based imputation methods | Using the SVD to perform ultra-efficient imputation | 2 min |
|
Probability densities on embeddings | Modeling distributions on data neighborhoods | 8 min |
|
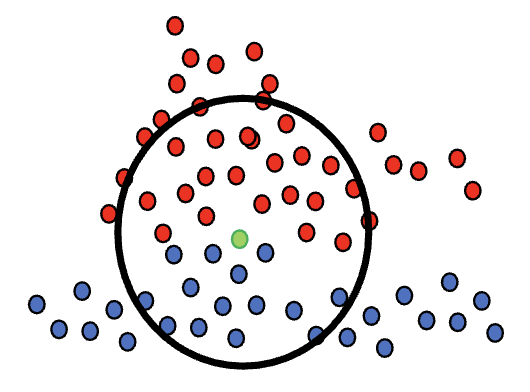
Which data are intrinsically hard to classify? | Margins for classifier embeddings | 13 min |

|
Evaluating extremely imbalanced classification | Finding needles in a haystack | 14 min |
|
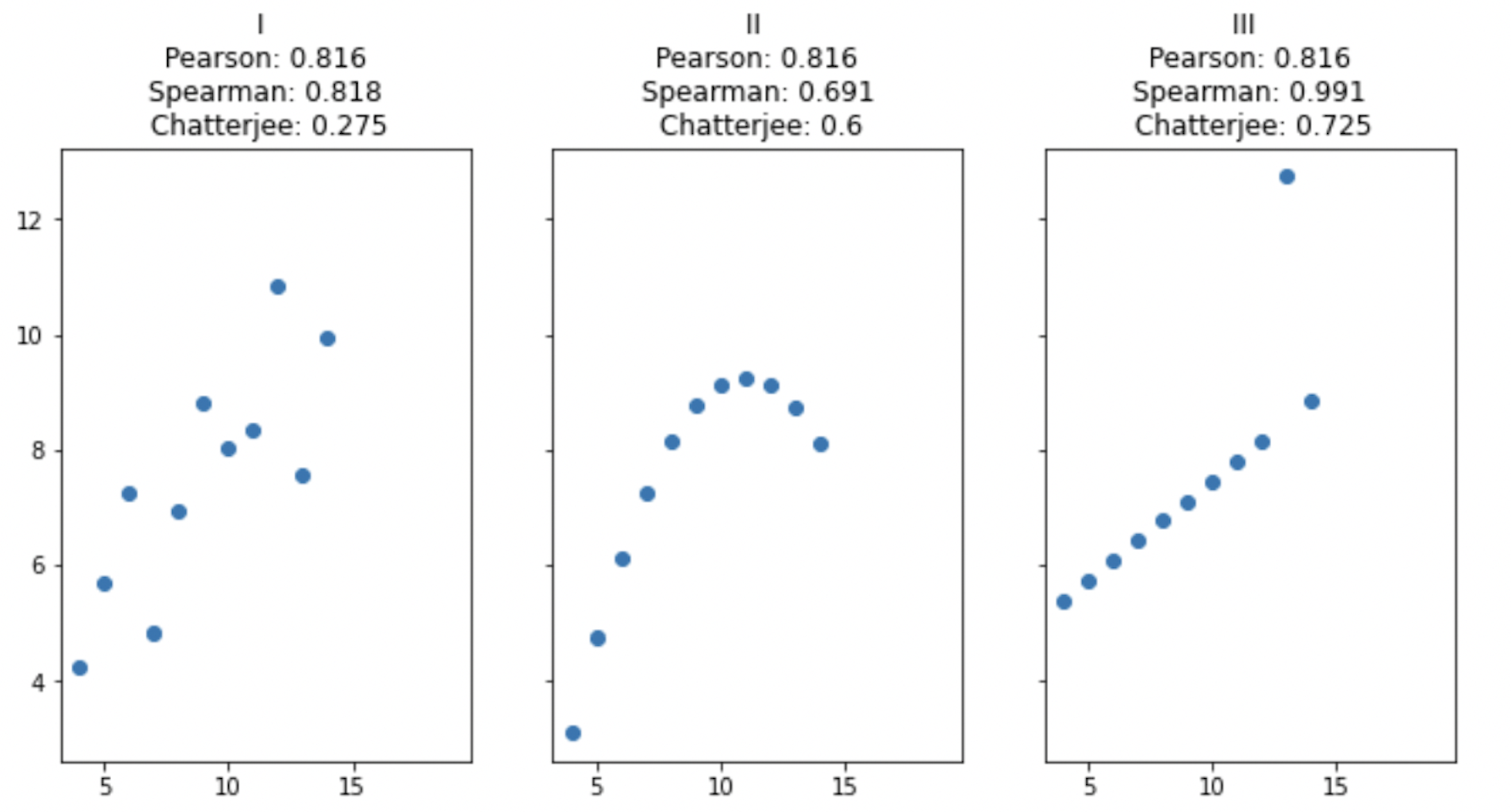
Measuring association between variables | A few new coefficients | 12 min |
|
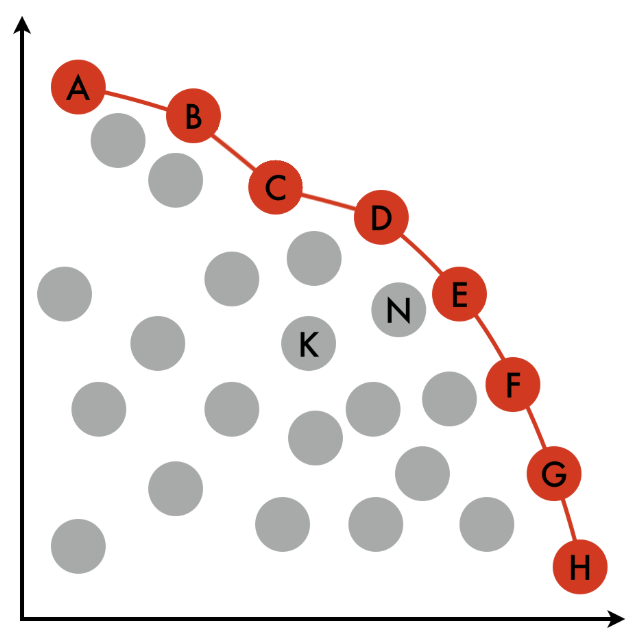
Multi-criterion optimization and decision-making | The power of weighted sums | 13 min |

|
Foundations of concentration and entropy | Where entropy comes from | 7 min |
No matching items